Rod Carty: DNA repair mechanisms make no sense in an evolutionary presupposition. Error correction requires error detection, and that requires the detection process to be able to compare the DNA as it is to the way it ought to be.
Kunkel, T.A., DNA Replication Fidelity, J. Biological Chemistry 279:16895–16898, 23 April 2004.
This machinery keeps the error rate down to less than one error per 100 million letters
Maintaining the genetic stability that an organism needs for its survival requires not only an extremely accurate mechanism for replicating DNA, but also mechanisms for repairing the many accidental lesions that occur continually in DNA.Most such spontaneous changes in DNA are temporary because they are immediately corrected by a set of processes that are collectively called DNA repair. Of the thousands of random changes created every day in the DNA of a human cell by heat, metabolic accidents, radiation of various sorts, and exposure to substances in the environment, only a few accumulate as mutations in the DNA sequence. For example, we now know that fewer than one in 1000 accidental base changes in DNA results in a permanent mutation; the rest are eliminated with remarkable efficiency by DNA repair. The importance of DNA repair is evident from the large investment that cells make in DNA repair enzymes. For example, analysis of the genomes of bacteria and yeasts has revealed that several percent of the coding capacity of these organisms is devoted solely to DNA repair functions.
Without DNA repair, spontaneous DNA damage would rapidly change DNA sequences
Although DNA is a highly stable material, as required for the storage of genetic information, it is a complex organic molecule that is susceptible, even under normal cell conditions, to spontaneous changes that would lead to mutations if left unrepaired.
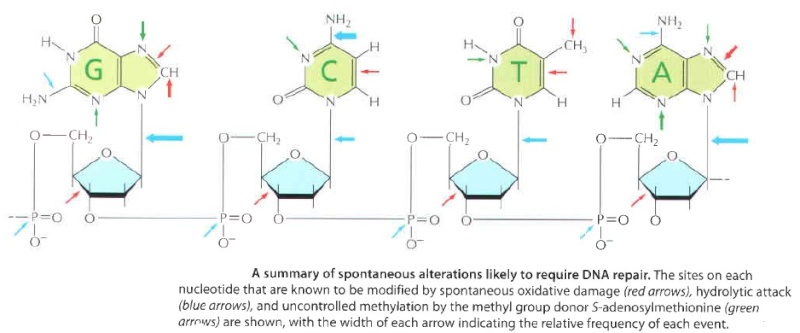
DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a base missing from the backbone of DNA, or a chemically changed base. 15
Naturally occurring DNA damages arise more than 60,000 times per day per mammalian cell.
DNA damage appears to be a fundamental problem for life. DNA damages are a major primary cause of cancer. DNA damages give rise to mutations and epimutations that, by a process of natural selection, can cause progression to cancer. 16
Different pathways to repair DNA
DNA repair mechanisms fall into 2 categories
– Repair of damaged bases
– Repair of incorrectly basepaired bases during replication
Cells have multiple pathways to repair their DNA using different enzymes that act upon different kinds of lesions.
At least four excision repair pathways exist to repair single stranded DNA damage:
Nucleotide excision repair (NER)
Base excision repair (BER)
DNA mismatch repair (MMR)
Repair through alkyltransferase-like proteins (ATLs)
In most cases, DNA repair is a multi-step process
– 1. An irregularity in DNA structure is detected
– 2. The abnormal DNA is removed
– 3. Normal DNA is synthesized
DNA bases are also occasionally damaged by an encounter with reactive metabolites produced in the cell (including reactive forms of oxygen) or by exposure to chemicals in the environment. Likewise, ultraviolet radiation from the sun can produce a covalent Iinkage between two adjacent pyrimidine bases in DNA to form, for example, thymine dimers This type of damage occurs in the DNA of cells exposed to ultraviolet or radiation(as in sunlight) A similar dimer will form between any two neighboring pyrimidine bases ( C or T residues ) in DNA. ( see below )
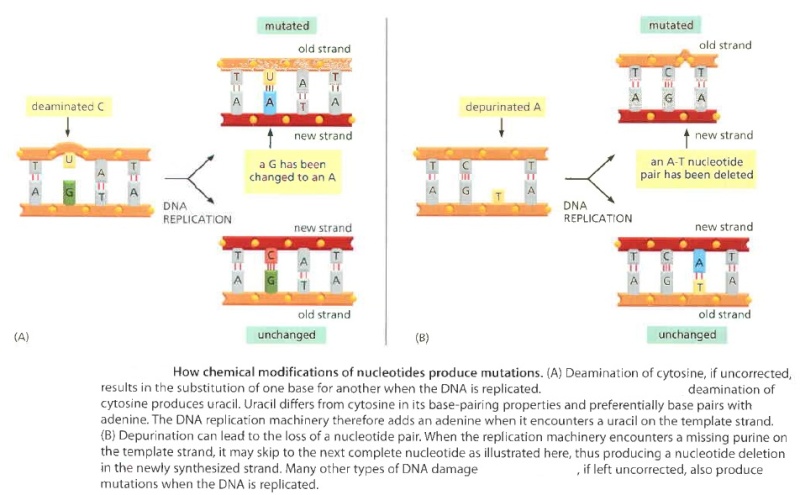
If left uncorrected when the DNA is replicated, most of these changes would be expected to lead either to the deletion of one or more base pairs or to a base-pair substitution in the daughter DNA chain. ( see below ) The mutations would then be propagated throughout subsequent cell generations. Such a high rate of random changes in the DNA sequence would have disastrous consequences for an organism
Its evident that the repair mechanism is essential for the cell to survive. It could not have evolved after life arose, but must have come into existence before. The mechanism is highly complex and elaborated, as consequence, the design inference is justified and seems to be the best way to explain its existence.
The DNA double helix is readily repaired
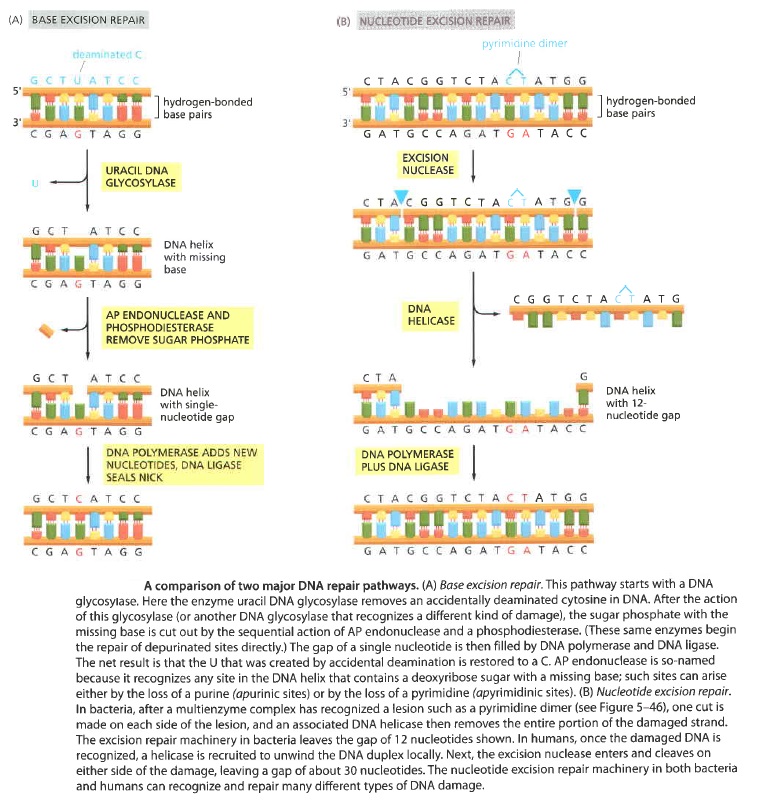
The double-helical structure of DNA is ideally suited for repair because it carries two separate copies of all the genetic information-one in each of its two strands. Thus, when one strand is damaged, the complementary strand retains an intact copy of the same information, and this copy is generally used to restore the correct nucleotide sequences to the damaged strand. An indication of the importance of a double-stranded helix to the safe storage of genetic information is that all cells use it; only a few small viruses use single stranded DNA or RNA as their genetic material. The types of repair processes described in this section cannot operate on such nucleic acids, and once damaged, the chance of a permanent nucleotide change occurring in these singlestranded genomes of viruses is thus very high. It seems that only organisms with tiny genomes (and therefore tiny targets for DNA damage) can afford to encode their genetic information in any molecule other than a DNA double helix.Below shows two of the most common pathways. In both, the damage is excised, the original DNA sequence is restored by a DNA polymerase that uses the undamaged strand as its template, and a remaining break in the double helix is sealed by DNA ligase.
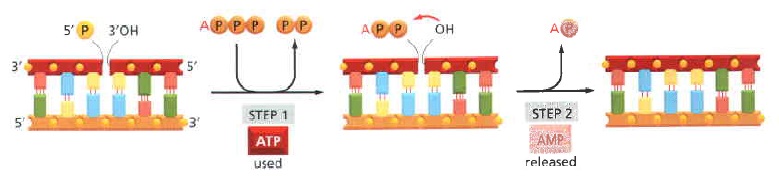
DNA ligase.
The reaction catalyzed by DNA ligase. This enzyme seals a broken phosphodiester bond. As shown, DNA ligase uses a molecule of ATP to activate the 5' end at the nick (step 1 ) before forming the new bond (step 2). In this way, the energetically unfavorable nick-sealing reaction is driven by being coupled to the energetically favorable
process of ATP hydrolysis.
The main two pathways differ in the way in which they remove the damage from DNA. The first pathway, called
Base excision repair (BER) 9
It involves a battery of enzymes called DNA glycosylases, each of which can recognize a specific tlpe of altered base in DNA and catalyze its hydrolltic removal. There are at least six types of these enzymes, including those that remove deaminated Cs, deaminated As, different types of alkylated or oxidized bases, bases with opened rings, and bases in which a carbon-carbon double bond has been accidentally converted to a carbon-carbon single bond.
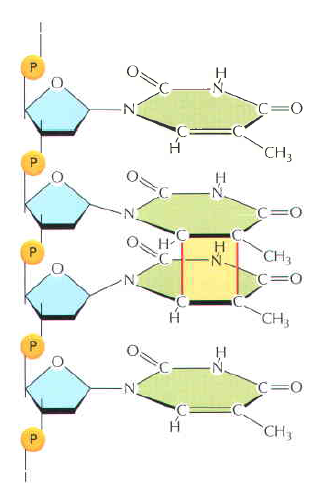
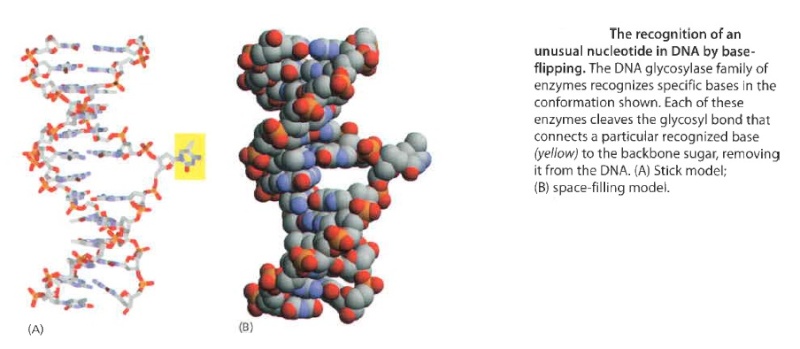
How is an altered base detected within the context of the double helix? A key step is an enzyme-mediated "flipping-out" of the altered nucleotide from the helix, which allows the DNA glycosylase to probe all faces of the base for damage ( see above image ) It is thought that these enzymes travel along DNA using base-flipping to evaluate the status of each base. Once an enzyme finds the damaged base that it recognizes, it removes the base from its sugar. The "missing tooth" created by DNA glycosylase action is recognized by an enzyme called AP endonuclease (AP for apurinic or apyrimidinic, endo to signify that the nuclease cleaves within the polynucleotide chain), which cuts the phosphodiester backbone, after which the damage is removed and the resulting gap repaired ( see figure below ) Depurination, which is by far the most frequent rype of damage suffered by DNA, also leaves a deoxyribose sugar with a missing base. Depurinations are directly repaired beginning with AP endonuclease.
While the BER pathway can recognize specific non-bulky lesions in DNA, it can correct only damaged bases that are removed by specific glycosylases. Similarly, the MMR pathway only targets mismatched Watson-Crick base pairs. 2
Molecular lesion A molecular lesion or point lesion is damage to the structure of a biological molecule such as DNA, enzymes, or proteins that results in reduction or absence of normal function or, in rare cases, the gain of a new function. Lesions in DNA consist of breaks and other changes in the chemical structure of the helix (see types of DNA lesions) while lesions in proteins consist of both broken bonds and improper folding of the amino acid chain. 6
DNA-N-glycosylases
Base excision repair (BER) involves a category of enzymes known as DNA-N-glycosylases These enzymes can recognize a single damaged base and cleave the bond between it and the sugar in the DNA removes one base, excises several around it, and replaces with several new bases using Pol adding to 3’ ends then ligase attaching to 5’ end
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond. Glycosylases were first discovered in bacteria, and have since been found in all kingdoms of life. 8
One example of DNA's automatic error-correction utilities are enough to stagger the imagination. There are dozens of repair mechanisms to shield our genetic code from damage; one of them was portrayed in Nature in terms that should inspire awe. 10
How do DNA-repair enzymes find aberrant nucleotides among the myriad of normal ones?
One enzyme has been caught in the act of checking for damage, providing clues to its quality-control process.
From Nature's article :
Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA 11
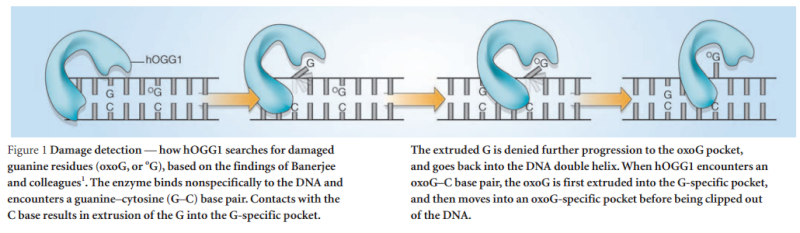
How DNA repair proteins distinguish between the rare sites of damage and the vast expanse of normal DNA is poorly understood. Recognizing the mutagenic lesion 8-oxoguanine (oxoG) represents an especially formidable challenge, because this oxidized nucleobase differs by only two atoms from its normal counterpart, guanine (G). The X-ray structure of the trapped complex features a target G nucleobase extruded from the DNA helix but denied insertion into the lesion recognition pocket of the enzyme. Free energy difference calculations show that both attractive and repulsive interactions have an important role in the preferential binding of oxoG compared with G to the active site. The structure reveals a remarkably effective gate-keeping strategy for lesion discrimination and suggests a mechanism for oxoG insertion into the hOGG1 active site.
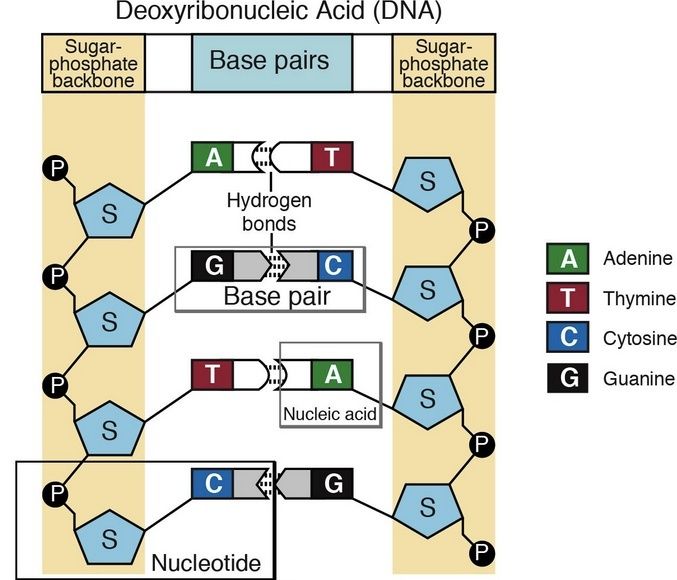
Of the four bases in DNA (C, G, A, and T) cytosine or C is always supposed to pair with guanine, G, and adenine, A, is always supposed to pair with thymine, T. The enzyme studied by Banerjee et al. in Nature is one of a host of molecular machines called BER glycosylases; this one is called human oxoG glycosylase repair enzyme (hOGG1), and it is specialized for finding a particular type of error: an oxidized G base (guanine). Oxidation damage can be caused by exposure to ionizing radiation (like sunburn) or free radicals roaming around in the cell nucleus. The normal G becomes oxoG, making it very slightly out of shape. There might be one in a million of these on a DNA strand. While it seems like a minor typo, it can actually cause the translation machinery to insert the wrong amino acid into a protein, with disastrous results, such as colorectal cancer. 12
The machine latches onto the DNA double helix and works its way down the strand, feeling every base on the way. As it proceeds, it kinks the DNA strand into a sharp angle. It is built to ignore the T and A bases, but whenever it feels a C, it knows there is supposed to be a G attached. The machine has precision contact points for C and G. When the C engages, the base paired to it is flipped up out of the helix into a slot inside the enzyme that is finely crafted to mate with a pure, clean G. If all is well, it flips the G back into the DNA helix and moves on. If the base is an oxoG, however, that base gets flipped into another slot further inside, where powerful forces yank the errant base out of the strand so that other machines can insert the correct one.
Now this is all wonderful stuff so far, but as with many things in living cells, the true wonder is in the details. The thermodynamic energy differences between G and oxoG are extremely slight – oxoG contains only one extra atom of oxygen – and yet this machine is able to discriminate between them to high levels of accuracy.
The author, David, says in the Nature article :
Structural biology: DNA search and rescue
DNA-repair enzymes amaze us with their ability to search through vast tracts of DNA to find subtle anomalies in the structure. The human repair enzyme 8-oxoguanine glycosylase (hOGG1) is particularly impressive in this regard because it efficiently removes 8-oxoguanine (oxoG), a damaged guanine (G) base containing an extra oxygen atom, and ignores undamaged bases.
The team led by Anirban Banerjee of Harvard, using a clever new stop-action method of imaging, caught this little enzyme in the act of binding to a bad guanine, helping scientists visualize how the machinery works. Some other amazing details are mentioned about this molecular proofreader. It checks every C-G pair, but slips right past the A-T pairs. The enzyme, “much like a train that stops only at certain locations,” pauses at each C and, better than any railcar conductor inspecting each ticket, flips up the G to validate it. Unless it conforms to the slot perfectly – even though G and oxoG differ in their match by only one hydrogen bond – it is ejected like a freeloader in a Pullman car and tossed out into the desert. David elaborates:
Calculations of differences in free energy indicate that both favourable and unfavourable interactions lead to preferential binding of oxoG over G in the oxoG-recognition pocket, and of G over oxoG in the alternative site. This structure [the image resolved by the scientific team] captures an intermediate that forms in the process of finding oxoG, and illustrates that the damaged base must pass through a series of ‘gates’, or checkpoints, within the enzyme; only oxoG satisfies the requirements for admission to the damage-specific pocket, where it will be clipped from the DNA. Other bases (C, A and T) may be rejected outright without extrusion from the helix because hOGG1 scrutinizes both bases in each pair, and only bases opposite a C will be examined more closely.
Natural selection cannot act without accurate replication, yet the protein machinery for the level of accuracy required is itself built by the very genetic code it is designed to protect. Thats a catch22 situation. It would have been challenging enough to explain accurate transcription and translation alone by natural means, but as consequence of UV radiation, it would have quickly been destroyed through accumulation of errors. So accurate replication and proofreading are required for the origin of life. How on earth could proofreading enzymes emerge, especially with this degree of fidelity, when they depend on the very information that they are designed to protect? Think about it.... This is one more prima facie example of chicken and egg situation. What is the alternative explanation to design ? Proofreading DNA by chance ? And a complex suite of translation machinery without a designer?
I enjoy to learn about the wonder of these incredible mechanisms. If the apostle Paul could understand that creation demands a Creator as he wrote in Romans chapter one 18, how much more we today with all the revelations about cell biology and molecular machines?
Since the editing machinery itself requires proper proofreading and editing during its manufacturing, how would the information for the machinery be transmitted accurately before the machinery was in place and working properly? Lest it be argued that the accuracy could be achieved stepwise through selection, note that a high degree of accuracy is needed to prevent ‘error catastrophe’ in the first place—from the accumulation of ‘noise’ in the form of junk proteins specified by the damaged DNA. 18
Depending on the species, this repair system can eliminate abnormal bases such as Uracil; Thymine dimers 3-methyladenine; 7-methylguanine
14
Since many mutations are deleterious, DNA repair systems are vital to the survival of all organisms
Living cells contain several DNA repair systems that can fix different type of DNA alterations
Nucleotide excision repair (NER)
Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals (i.e. intercalating agents), radiation and other mutagens.
Nucleotide excision repair (NER) is a highly conserved DNA repair mechanism. NER systems recognize the damaged DNA strand, cleave it on both sides of the lesion, remove and newly synthesize the fragment. UvrB is a central component of the bacterial NER system participating in damage recognition, strand excision and repair synthesis.[/b] We have solved the crystal structure of UvrB in the apo and the ATP-bound forms. UvrB contains two domains related in structure to helicases, and two additional domains unique to repair proteins. The structure contains all elements of an intact helicase, and is evidence that UvrB utilizes ATP hydrolysis to move along the DNA to probe for damage. The location of conserved residues and structural comparisons allow us to predict the path of the DNA and suggest that the tight preincision complex of UvrB and the damaged DNA is formed by insertion of a flexible β-hairpin between the two DNA strands. 3
DNA constantly requires repair due to damage that can occur to bases from a vast variety of sources including chemicals but also ultraviolet (UV) light from the sun. Nucleotide excision repair (NER) is a particularly important mechanism by which the cell can prevent unwanted mutations by removing the vast majority of UV-induced DNA damage (mostly in the form of thymine dimers and 6-4-photoproducts). The importance of this repair mechanism is evidenced by the severe human diseases that result from in-born genetic mutations of NER proteins including Xeroderma pigmentosum and Cockayne's syndrome. While the base excision repair machinery can recognize specific lesions in the DNA and can correct only damaged bases that can be removed by a specific glycosylase, the nucleotide excision repair enzymes recognize bulky distortions in the shape of the DNA double helix. Recognition of these distortions leads to the removal of a short single-stranded DNA segment that includes the lesion, creating a single-strand gap in the DNA, which is subsequently filled in by DNA polymerase, which uses the undamaged strand as a template. NER can be divided into two subpathways (Global genomic NER and Transcription coupled NER) that differ only in their recognition of helix-distorting DNA damage. 4
Nucleotide excision repair (NER) is a particularly important excision mechanism that removes DNA damage induced by ultraviolet light (UV). 2UV DNA damage results in bulky DNA adducts - these adducts are mostly thymine dimers and 6,4-photoproducts. Recognition of the damage leads to removal of a short single-stranded DNA segment that contains the lesion. The undamaged single-stranded DNA remains and DNA polymerase uses it as a template to synthesize a short complementary sequence. Final ligation to complete NER and form a double stranded DNA is carried out by DNA ligase. NER can be divided into two subpathways: global genomic NER (GG-NER) and transcription coupled NER (TC-NER). The two subpathways differ in how they recognize DNA damage but they share the same process for lesion incision, repair, and ligation.
The importance of NER is evidenced by the severe human diseases that result from in-born genetic mutations of NER proteins. Xeroderma pigmentosum and Cockayne's syndrome are two examples of NER associated diseases.
Maintaining genomic integrity is essential for living organisms. NER is a major pathway allowing the removal of lesions which would otherwise accumulate and endanger the health of the affected organism. 5
Nucleotide excision repair (NER) is a mechanism to recognize and repair bulky DNA damage caused by compounds, environmental carcinogens, and exposure to UV-light. In humans hereditary defects in the NER pathway are linked to at least three diseases: xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy (TTD). The repair of damaged DNA involves at least 30 polypeptides within two different sub-pathways of NER known as transcription-coupled repair (TCR-NER) and global genome repair (GGR-NER). TCR refers to the expedited repair of lesions located in the actively transcribed strand of genes by RNA polymerase II (RNAP II). In GGR-NER the first step of damage recognition involves XPC-hHR23B complex together with XPE complex (in prokaryotes, uvrAB complex). The following steps of GGR-NER and TCR-NER are similar.
1) http://www.genome.jp/dbget-bin/www_bget?ko03420
2) http://en.wikipedia.org/wiki/Nucleotide_excision_repair
3) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1171753/pdf/006899.pdf
4) http://bioisolutions.blogspot.com.br/2008/04/ner-pathway.html
5) http://intelligent-sequences.blogspot.com.br/2008_06_01_archive.html
6) https://en.wikipedia.org/wiki/Molecular_lesion
8 ) https://en.wikipedia.org/wiki/DNA_glycosylase
9) http://www.csun.edu/~cmalone/pdf360/Ch15-2repairtanspose.pdf
10) http://www.nature.com/nature/journal/v434/n7033/full/nature03458.html
11) http://www.nature.com/nature/journal/v434/n7033/full/nature03458.html
12) http://creationsafaris.com/crev200503.htm
13) http://fire.biol.wwu.edu/trent/trent/DNAsearchrescue.pdf
14) http://www.genome.jp/kegg-bin/show_pathway?ko03410
15) https://en.wikipedia.org/wiki/DNA_damage_(naturally_occurring)
16) http://www.intechopen.com/books/new-research-directions-in-dna-repair/dna-damage-dna-repair-and-cancer
17) http://creation.com/dna-best-information-storage


Nenhum comentário:
Postar um comentário